Research
Transposable elements (TEs) are mobile genetic sequences that populate and parasitize genomes. TEs impose a multifaceted mutational burden on their host through insertional inactivation of host genes, participation in ectopic recombination, and the induction of DNA damage. Our research focuses on how the host genome responds fight back against TEs through small RNA silencing pahtways, and how they tolerate TE activity in developing germline cells.
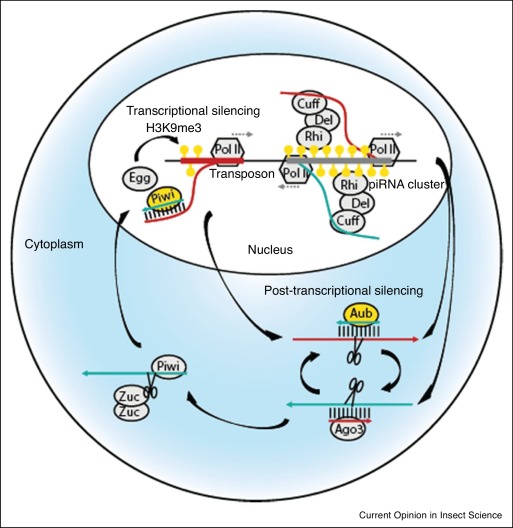
Schematic of piRNA mediated regulation of TEs (from Kelleher, Lama and Wang, 2020)
Resistance
Evolution of small RNA mediated silencing
Eukaryotic genomes defend themselves from TEs through small RNA-mediated silencing pathways, which supress the transcription and translation of TE encoded proteins required for transposition. We study how these pathways evolve.
More Info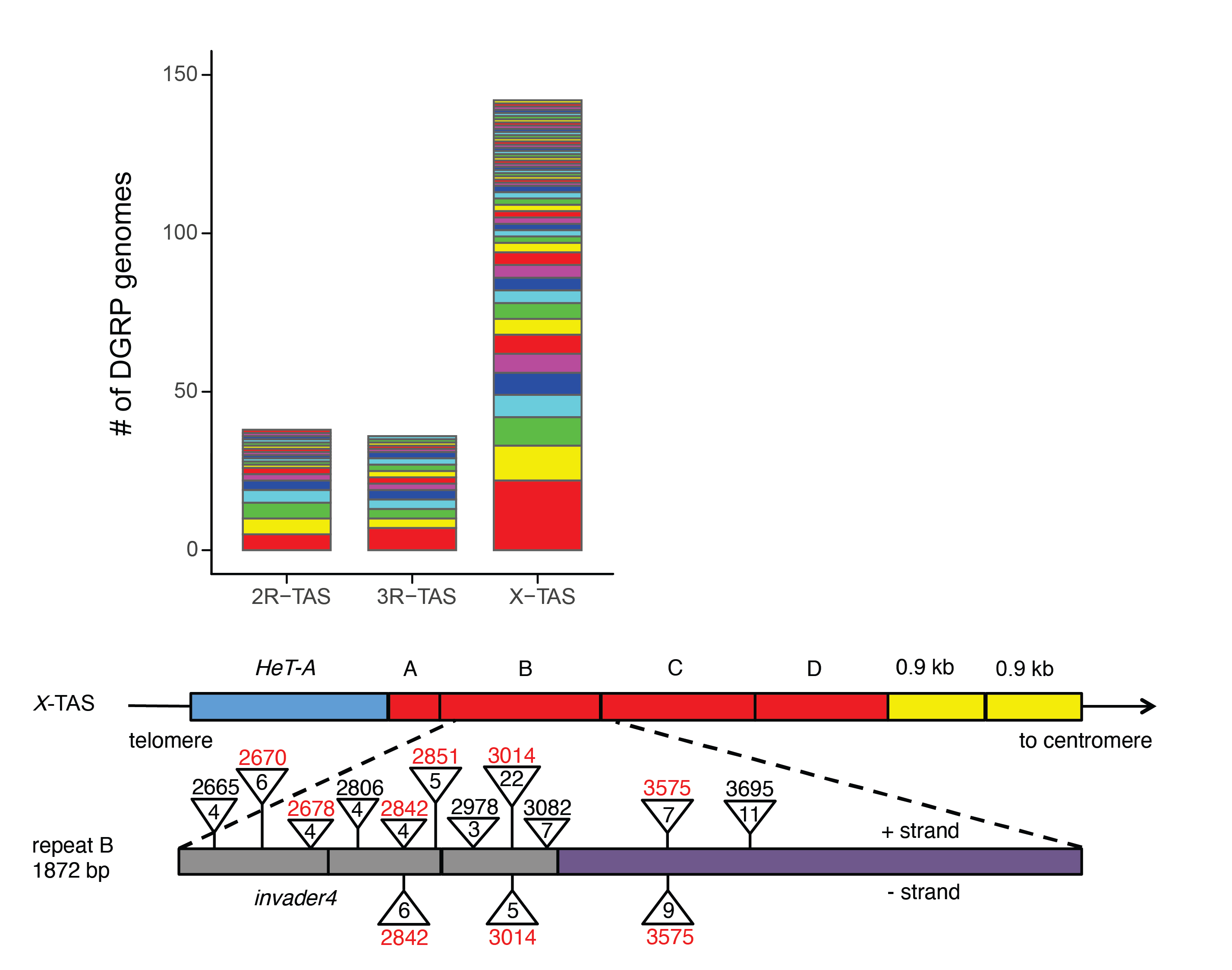
Hundreds of P-element insertions into TAS piRNA clusters arose over the course of 50 years in D. melanogaster, allowing for the rapid evolution of resistance (from Zhang, Pointer and Kelleher 2020).
Evolved resistance of invading TEs.
When new TEs invade the genome, how are new species of piRNAs produced? We took advantage of the recent invasion of the Drosophila melanogaster genome by P-element DNA transposons to reconstruct the mutations that brought the TE under host control. By contrasting historic and recent collections of flies that differ in their ability to repress P-elements, we discovered that piRNA mediated silencing evolved rapidly through frequent insertion of P-elements into piRNA producing sites. We are currently studying the evolution of resistance in laboratory populations, in order to draw closer connections between transposition into piRNA clusters and piRNA-mediated immunity. We are also interested in the role of epigenetic mutations in creating new piRNA-producing alleles.
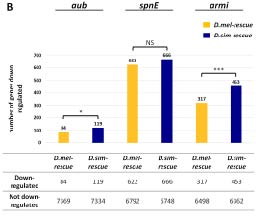
Heterospecific alleles of adaptively evolving piRNA pathway proteins exhibit enhanced negative regulation of host genes, suggesting increased genomic autoimmunity (from Wang, Barbash and Kelleher 2020).
Genomic Autoimmunity
While the piRNA pathway’s role in TE regulation is critical for genome integrity, how does it avoid off-target silencing of host genes. Our recent work that suggests among adaptively evolving cytoplasmic piRNA proteins, avoidance of interactionw ith host transcripts is an important target of selection. In the future, we hope to better understand the contribution of off-target effects on host gene regulation.
P-elemement transposition leads to a loss of germline cells (From Kelleher et al. 2018)
Tolerance
How do germline cells withstand the genotoxic effects of TEs?
We are also interested in how host cells, particularly developing germline cells, withstand the genotoxic effects of TEs. DNA damage resulting from transposition is associated with the loss and disrupted development of male and female gametes. Are there existing mechanisms that allow host cells to tolerate germline DNA damage, thereby ensuring host fertility?
More Info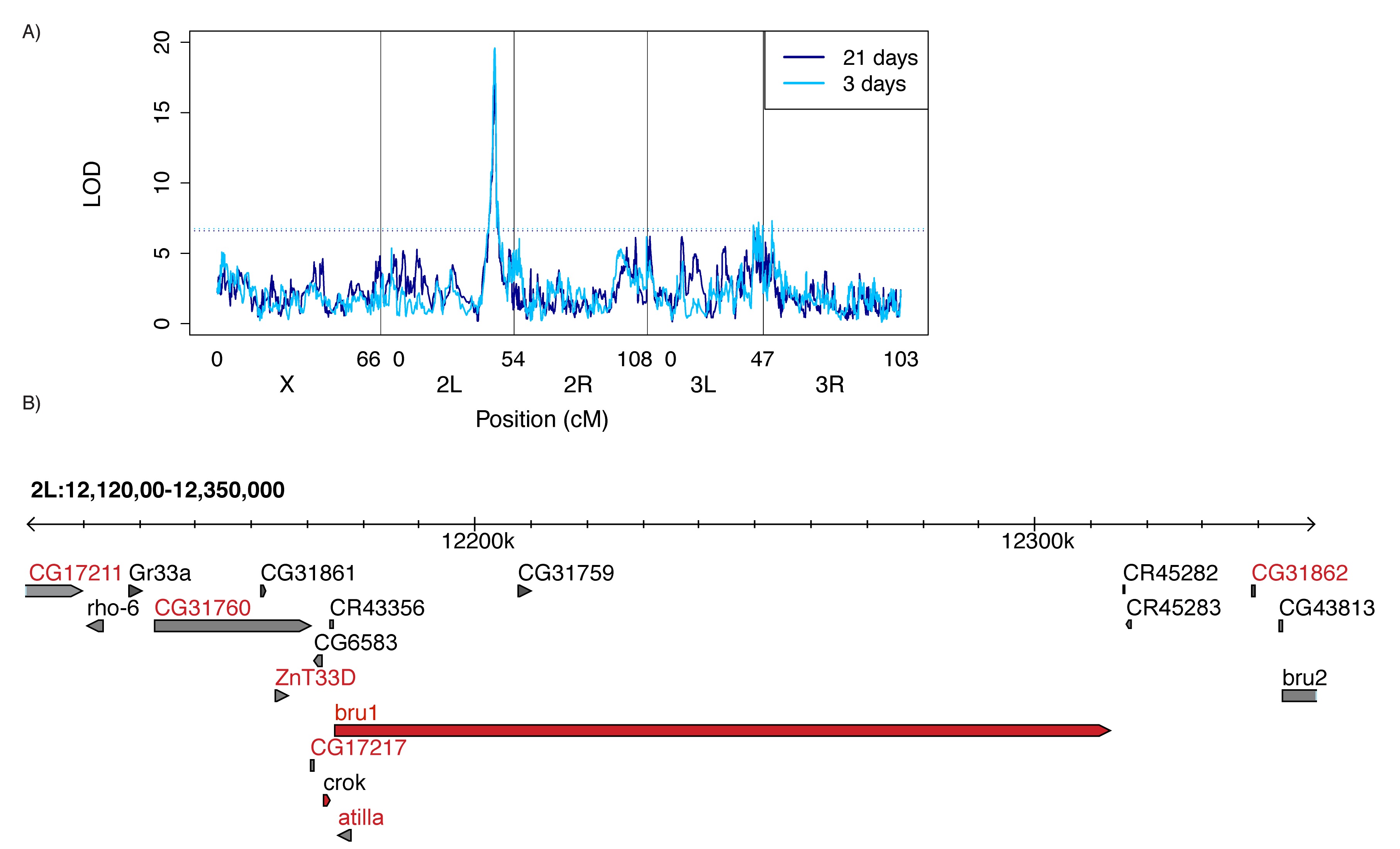
QTL mapping of natural variation in tolerance revealed that reduced bruno expression is associated with reduced P-element induced sterility, the first example of a tolerant allele in any organism (from Kelleher et al 2018).
Natural variation in tolerance
Through QTL mapping of recombinant inbred lines we have uncovered multiple genomic regions that display natural variation in tolerance to P-transposable elements. Flies carrying tolerant alleles produce gametes even in the absence of piRNA-mediated regulation, while individuals sensitive alleles are sterile. Our current work focuses on understanding the cellular mechanisms of tolerance, as well as the recent evolutionary history of tolerance alleles.
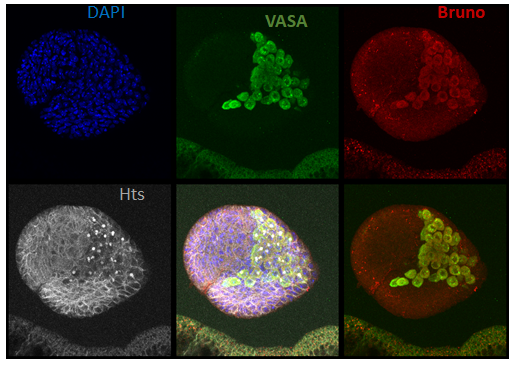
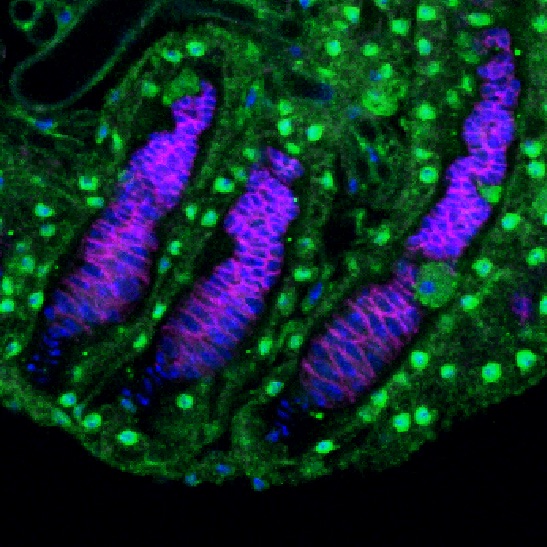
Bruno protein expression in primordial germ cells of a stage-3 female larvae (photo from Modupeola Bolaji).
Mechanism of bruno-dependent tolerance
Bruno is an RNA binding protein with known functions in the regulation of oogenesis in adult females. We have recently discovered that bruno mutants suppress the loss of primordial germ cells (PGCs) in the ovaries of dysgenic females, and that Bruno protein is expressed in PGCs (left). We are currently exploring how bruno function may impact the regulation of P-elements in PGCs, or their response to transposition.