

| Home | Research | Publications | Lab info | Links | Contact us | Protocols |
|---|
LincRNAs and Stem Cells Our research is centered on stem cell
differentiation of the cardiac lineage and heart development. One of our goals
for this research is to characterize the epigenetic and transcriptome changes
that occur during early embryonic heart differentiation. Specifically, We are interested in the
epigenetic modifications involving long, intergenic non-coding RNAs and their
role in defining the cardiac lineage. Recently, lincRNAs have emerged as one of
the most important regulatory paradigms in mammalian biology by partnering with
genomic enhancer regions and thereby co-activating and/or co-repressing transcriptional
activity of the targeted genomic region. It is plausible that lincRNAs can
regulate histone “readers,” “writers,” or “erasers” allosterically altering
their ability to modulate repressive or enhancive epigenetic marks.
|
 |
|---|---|
Transcriptional and Epigenetic regulation of cardiac Stem Cells We are interested in transcriptional and epigenetic regulation of cardiac
stem cells in the context of vertebrate embryogenesis causing human
congenital heart defects (CHDs), and their implications in adult
cardiac muscle regeneration. In particular, We are interested in
determining the roles of transcription factors and molecular signalling
pathways in cardiac lineage specification, including T-box
transcription factors, Wnt signaling, Notch signaling, and the role of
epigenetic chromatin remodeling complexes including Polycomb Repressive
Complex 2 (PRC2) and BAF complex. Our goal is to obtain an in-depth
understanding of these molecular pathways in order to prevent neonatal
CHDs and efficiently regenerate heart muscle in adult patients with
irrevocable heart diseases. | 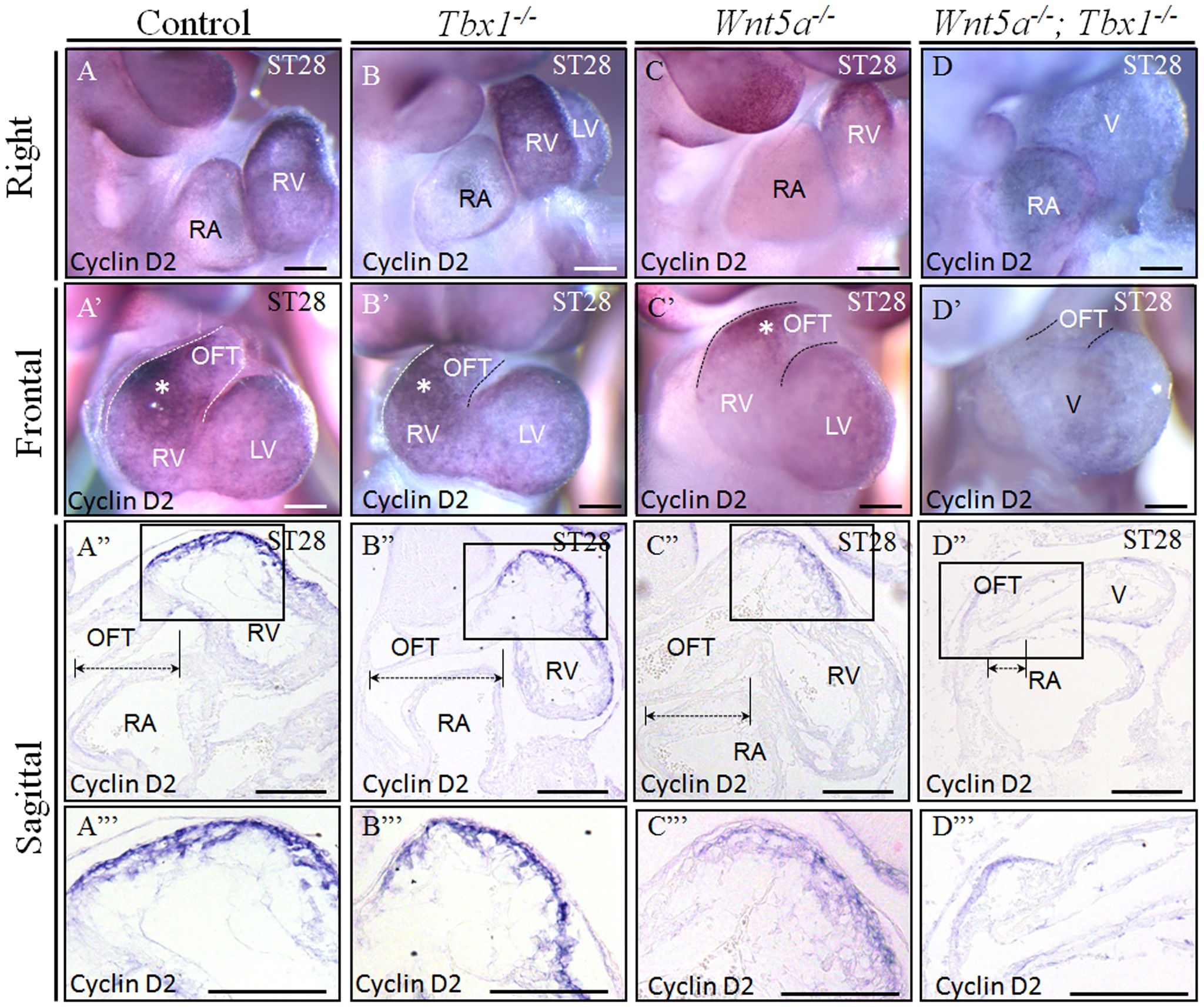 |
Molecular regulation of Heart development | |
Cardiac progenitor cells (CPCs) We are interested in the fate commitment and
maturation of cardiac progenitor cells (CPCs). CPCs hold a unique position in
heart regenerative medicine for a number of reasons. First, they can
differentiate into multiple cardiovascular lineages, including smooth muscle,
endothelial, and cardiac muscle for a concerted therapeutic effect. Second, unlike totipotent ES cells, they do
not form tumor when grafted. Third, unlike terminally differentiated cells,
they are proliferative thus a relatively small amount may be sufficient for
therapeutic effects. We are studying what extracellular cues induce CPCs and
what signaling pathways drive the differentiation of them into the major
cardiovascular lineages. In collaboration with other labs in the college, we
are exploring the opportunity to use CPCs for experimental therapy in moue
heart disease models. Another related research interest is the function of
endoderm in cardiac differentiation. Previously, We have found that the endoderm
protein Sox17 has a pivotal role in inducing CPCs. We are delinearizing this
pathway, in hope to ultimately find the secreted factor mediating the induction
of CPCs. |  |