|
Parallel synthesis of combinatorial libraries of molecules is an evolving field at the interface of chemistry, medicinal chemistry, biology, and microengineering. Combinatorial synthesis is to prepare a large number of molecules (library molecules) in one experiment. There are several methods for combinatorial synthesis, which differ in the way of handling library molecules, scale (number of molecules synthesized), product quantity, efficiency, quality, and many other factors (Table 1). The synthesis method #3 in Table 1 was developed in this laboratory in collaboration with Dr. Xiaochuan Zhou (Atactic Technologies Inc. Houston) and Dr. Erdogan Gulari (University of Michigan) and their research teams. An example of parallel synthesis of addressable microarrays (addressable: the identity of the compound at each location of the surface is known) is illustrated below (Figure 2). Comparing to conventional reactions, parallel synthesis relies on two key processes:
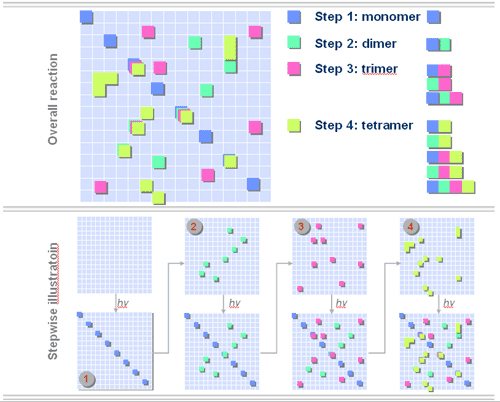
Figure 2. Illustration of a four step parallel synthesis on solid surface. Color squares represent different chemical building block monomers for library synthesis. Step 1 is the first cycle of the synthesis which involves a unique light gating pattern and adding of the first building block monomer. Table 1. Combinatorial Synthesis Methods – Select
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| UofH | Chemistry | Biology&Biochemistry | ACS | NCBI | PDB | CSCPDB | SeqTools | NMRTools Login | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||