|
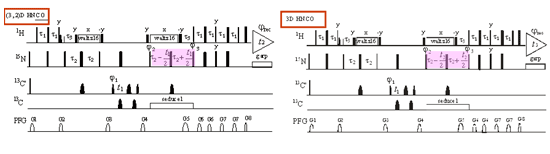
While NMR is clearly the only solution-based method for high resolution structures of large molecules, but the technique is not widely adopted in research laboratories. The barriers to wide scale use mainly reside in a steep learning curve in NMR experiments, spectral interpretation, and structure computation. The extensive effort required to solve a NMR structure and the lengthy amount of time of months or even years also prohibits NMR keeping in pace with rapidly evolving biological research. To address these issues, we are developing a suite of NMR methods to put NMR in the fast lane. Our efforts are in two areas: fast NMR experiments and fast NMR spectral interpretation. (a) Fast NMR experiments: Our lab applies two principal methods in new fast NMR experiments, one is time sharing data acquisition scheme (TS-NMR) and the other is frequency editing in reduced dimensions during data collection/frequency deconvolution during data processing (GFT-NMR). We developed TS (C,N)-HSQC and (C,N)-NOESY-HSQC experiments and various GFT-NMR experiments, achieving reduction of NMR time by multiple folds without sacrificing spectral quality. A representative pulse sequence of GFT (3,2)D HNCO (2D data for 3D H-N-CO connectivities and frequency editing of N, CO in the indirect dimension) is shown in Figure 10. We have developed a suite of GFT-NMR experiments and data processing tools to allow complete 3D data set collection in 24 hours. Furthermore, we demonstrate TS- and GFT-NMR data acquisition methods can be used in combination to achieve high resolution NOE detection in a much shorter amount of time and at a much higher spectral resolution.
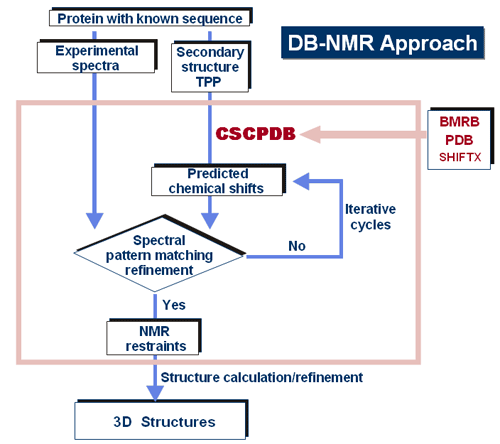
Figure 10. Pulse scheme of GFT (3,2)D HNCO. This experiment generates 2D data for 3D H-N-CO connectivities based on frequency editing of N, CO in the indirect dimension (see shaded evolution time). The experiment takes 10 min and is 18-times shorter than an equivalent conventional 3D HNCO experiment. A requirement for these new, advanced NMR experiments to be of general value is to restore the frequency sharing or convoluted data to conventional spectral appearance. For these applications, we have modified SPARKY and created data processing to facilitate data interpretation. (b) Fast spectral interpretation methods: Despite of many years of effort in automation of NMR spectral interpretation, this goal is still beyond reach. Given the amount of the available chemical shift assignments and structures and reasonable theoretical predictions of chemical shifts from known 3D structures (PDB databank), we have constructed a comprehensive chemical shift correlated protein database (CSCPDB) in which chemical shifts of 1H, 13C, and 15N are annotated according to their residue identity, sequence, and structure for over 2,000 proteins. This database is of great value for examining structure and sequence correlations of peptide sequences [link], and it can be used to ôsynthesize?NMR spectra for comparison purposes. Using the millions of chemical shift data points in CSCPDB as the training set, we are developing a database-driven method in combination with conventional NMR assignment rules for complete automation of NMR spectral interpretation for proteins. The basics of our project is shown in Figure 11.
Figure 11. Database driven approach for complete automation of NMR spectra Ultimately, a non-expert user program will be developed to allow NMR spectral analysis in a matter of a few hours or minutes. The DB-NMR method will be web-interactive and accessible.
|
||
| UofH | Chemistry | Biology&Biochemistry | ACS | NCBI | PDB | CSCPDB | SeqTools | NMRTools Login | ||