|
In the last twenty years, nuclear magnetic resonance (NMR) spectroscopy continues to evolve as a powerful technology for observing molecular details in solution. The importance of NMR is evident for Nobel Prizes awarded to leading researchers in the field, to Drs. Richard Ernest (1991), Kurt WŁthrich (2002), and Paul C. Lauterbur (2003). The University of Houston has established high resolution NMR Center since 1993 (the Keck/IMD NMR Center). We have direct access to the state-of-the-art 800 MHz (Figure 8) and 600 MHz NMR instruments on the campus.
Figure 8. A view of the Keck/IMD NMR Center at the University of Houston. The newly renovated facility of 8,000 ft2 houses the 800 MHz and the 600 MHz instruments, wet-labs, and offices. The required electrical power, ultra-dry air compressor, process chiller, and other necessary accessories for the requested CryoProbe systems have been installed. Our group has solved novel structures of ligand-DNA complexes, disease-related DNA molecules, drug-like DNA and RNA molecules, and proteins (Table 2). A typical NMR spectrum of a 230 residue protein is shown in Figure 9.
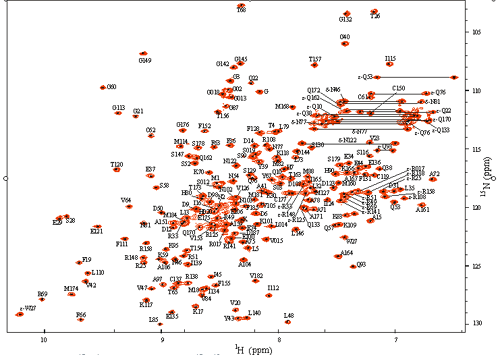
Figure 9. 2D 15N-1H HSQC of uniformly 15N/13C-labled CUGBP1. Assigned residues are marked with their residue name and residue number. In our effort of NMR for structure and function studies, a list of our interested projects is as following:
The second portion of our NMR effort is method development. Dr. Youlin Xia is leading this effort in creating new NMR experiments to overcome the problems associated with overcrowded spectra, low signal sensitivity, and increasingly large proteins in high resolution NMR studies. The new NMR experiments and related software tools for data process and spectral analysis can be found at our links of tools.
|
||
| UofH | Chemistry | Biology&Biochemistry | ACS | NCBI | PDB | CSCPDB | SeqTools | NMRTools Login | ||